
| Layer group number | 69 |
| Bilayer ID | BaCl2-00a09165a480-2-1_-1_1_0--0.33_-0.67 |
| Slide Stability | - |
| Space group | P3m1 |
| Binding Energy (gs) [meV/Å2] | 13.500 |
| Distance [Å] | 2.306 |
| Space group number | 156 |
| Band gap (PBE) | 5.300 |
| Magnetic | No |
| Layer group | p3m1 |
| Number of layers | 2 |
| Monolayer ID | BaCl2-00a09165a480 |
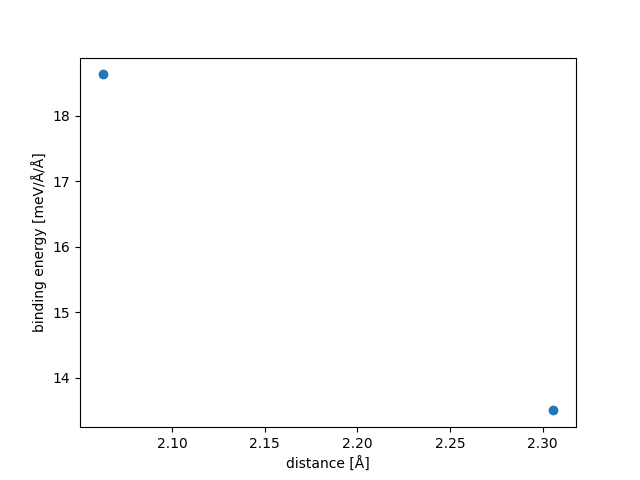
| Stacking | Binding energy [meV/Å2] | Slide Stability |
|---|---|---|
| 1BaCl2-1-2-1_-1_1_0--0.33_-0.67 | 13.500 | - |
| 1BaCl2-1-2-1_0_0_1--0.33_-0.67 | 18.635 | Stable |
| Key values [eV] | |
|---|---|

| Miscellaneous details | |
|---|---|
| Unique ID | BaCl2-00a09165a480-2-1_-1_1_0--0.33_-0.67 |
| Number of atoms | 6 |
| Number of species | 2 |
| Formula | Ba2Cl4 |
| Reduced formula | BaCl2 |
| Stoichiometry | AB2 |
| Unit cell area [Å2] | 20.066 |
| evac | 3.442 |
| folder | /home/niflheim2/cmr/WIP/stacking/tree-mads/tree/AB2/BaCl2/BaCl2-00a09165a480/BaCl2-2-1_-1_1_0--0.33_-0.67 |
| Monolayer ID | BaCl2-00a09165a480 |
| Bilayer ID | BaCl2-00a09165a480-2-1_-1_1_0--0.33_-0.67 |
| Number of layers | 2 |
| Layer group number | 69 |
| Layer group | p3m1 |
| Space group number | 156 |
| Miscellaneous details | |
|---|---|
| Space group | P3m1 |
| equivalent_atoms | [0 1 2 3 4 5] |
| has_inversion_symmetry | No |
| Magnetic | No |
| Band gap (PBE) | 5.300 |
| gap_pbe_dir | 5.338 |
| gap_pbe_nosoc | 5.317 |
| gap_pbe_dir_nosoc | 5.357 |
| dipz | -0.039 |
| with_props | No |
| Slide Stability | - |
| Binding energy (zscan) | 13.415 |
| Binding Energy (gs) [meV/Å2] | 13.500 |
| Distance [Å] | 2.306 |
